The cost of next generation sequencing (NGS) has been steadily decreasing with the invention of new technologies and improved chemistry of sequencing devices.1 As operational costs for NGS decrease, its relevance is soaring, extending not only to direct-to-consumer applications but also encompassing certified service providers and a growing number of clinicians who are harnessing NGS for disease diagnosis and personalized medicine. Numerous case reports have highlighted the effectiveness of NGS in identifying a wide array of pathogens, including viruses, bacteria, fungi, and parasites, through the analysis of microbial samples.2
As the usage of NGS continues to increase, so too does the demand for high-throughput extraction kits. This reliance on high-throughput extraction methods increases the probability of sample well-to-well cross-contamination occurring in 96-well racks.3 While contamination can have various sources, including the laboratory environment, researchers, plastic consumables, and nucleic acid extraction kits4, 96-well lysis racks have been pinpointed as a frequent source of leakage and cross-contamination during mechanical lysis. Mechanical lysis stands out for its precision in determining microbial diversity, by enabling extraction of difficult to lyse microorganisms5, which sets it apart from less robust alternatives such as chemical lysis.
This study aims to provide a comparison of commercially available 96-well lysis racks. Identifying potential factors contributing to leakage or cross-contamination can help researchers, clinicians, and laboratories avoid costly errors in disease diagnosis and quantifying microbial communities. Leakage, characterized by the unintentional escape of a substance due to containment failure, and cross-contamination, involving the transfer of substances between surfaces, represent two distinct yet interconnected challenges. Instances of sample leakage carry the risk of cross contaminating other components in a workflow, resulting in failed outcomes. Leakage and cross-contamination not only lead to sample volume loss but also introduce significant biohazard risk, unless all samples are adequately preserved using an inactivation solution like DNA/RNA ShieldTM. Both leakage and cross-contamination pose challenges to a workflow and can have serious downstream consequences. Herein Zymo Research investigates the incidence of leakage and well-to-well cross-contamination of commercially available 96-well lysis racks with a specific focus on the lysis stage of high throughput extractions.
Assessing Cross-Contamination and Leakage
The basic principle behind this experimental design was creating a simple and elegant checkered color scheme in a 96-well lysis rack using food dye. Experimental titrations determined that as little as one microliter of food dye infiltrating any adjacent wells would create a noticeable color change. Figure 1, provided below, is a visual representation of the designed 96-well checkered color scheme. Colored wells utilize undiluted food coloring (red or blue in color) and DNA/RNA ShieldTM, while non-colored wells contained only the DNA/RNA ShieldTM reagent.
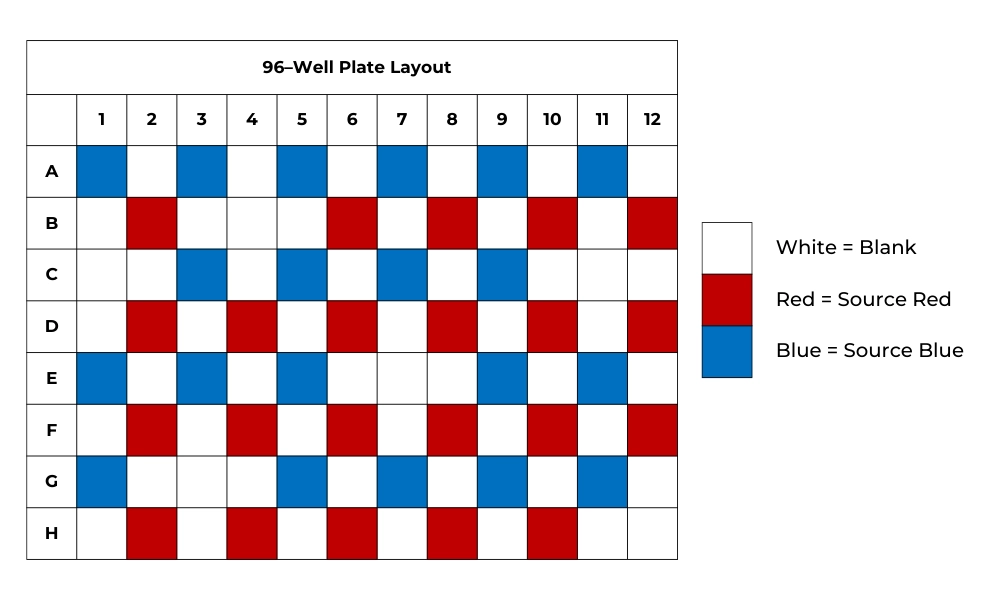
Figure 1: 96-Well Lysis Plate Setup. The setup plate is how an individual 96-well lysis rack was loaded onto each machine.
Each plate type was tested on two different 96-well mechanical lysis machines. The Benchmark BeadBlasterTM 96 Ball Mill Homogenizer is a mechanical homogenizer with a two-dimensional shaking action that moves the lysis racks in an X and Y axis. In contrast, the BioSpec Mini-BeadBeater-96 is a mechanical homogenizer with a three-dimensional shaking action that moves lysis racks in the X, Y and Z axis; this results in an overall circular motion. Figure 2 below depicts the motion of the Benchmark BeadBlasterTM 96 Ball Mill Homogenizer and the BioSpec Mini-BeadBeater-96. Both of these devices are widely used in the field of microbiomics and were selected to be representative of the motion of similar homogenizer’s whether unidirectional or multidirectional.
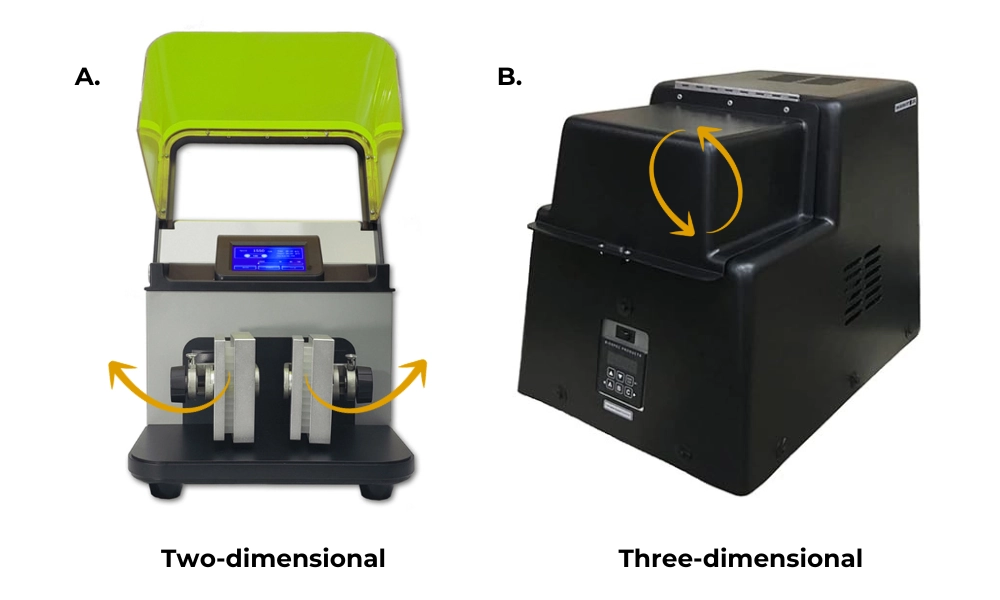
Figure 2: Direction of the mechanism. Benchmark BeadBlasterTM 96 Ball Mill Homogenizer (A) has motion in two dimensions, the X and Y axis. The BioSpec Mini-Beadbeater-96 (B) has motion in three dimensions, the X, Y and Z axis, resulting in a circular motion.
Each 96-well lysis rack was secured with the closure recommended per the manufacturer’s instructions (i.e., closure mat, PCR-grade seal, strip tube caps, screw caps).Table 1, presented below, provides an overview of each commercially available 96-well rack and its associated closure. Each lysis rack underwent five minutes of shaking, followed by a five-minute rest period, for a maximum of eight cycles. Before and after each lysing event, photos were taken from a top-down view of each individual 96-well plate to observe the extent of leakage or cross-contamination (see supplementary images).
|
96-Well Lysis Rack |
Closure Provided |
Sealing Method |
|---|---|---|
|
Qiagen PowerBead Pro Plate |
PCR Seal |
Adhesive |
|
MP Bio Lysing Matrix B, 96-tube Rack |
Strip Tube Caps |
Press Fit |
|
Omni 96 Deep Well Plate |
Silicon Mat |
Press Fit |
|
Applied BiosystemsTM MagMAXTM Microbiome Bead Plate |
PCR Seal & 2 Foil Seals |
Adhesive |
|
BioSpec 2ml Deep Well Microplate |
Silicon Mat |
Press Fit |
|
ZR-96 BashingBeadTM Lysis Rack (Barcoded) |
Plastic |
Threaded Screw Cap |
|
ZymoBIOMICS BashingBeadTM Lysis Rack (0.5mm & 0.1mm) |
Foil Seal |
Heat Bonded |
Which Racks Are Prone to Cross-Contamination and Leakage?
Our study investigated seven commercially available 96-well lysis racks. Among these, five of seven (5/7) lysis racks displayed both leakage and cross-contamination on the BioSpec Mini-BeadBeater-96. Similarly, on the Benchmark BeadBlasterTM 96 Ball Mill Homogenizer four of seven (4/7) racks showed leakage and cross-contamination. Notably, the Omni 96 Deep Well plate showed leakage but no visible colored cross-contamination on the Benchmark machine. Table 2 presents a comprehensive account of the data observed during testing with the Benchmark BeadBlasterTM 96 Ball Mill Homogenizer. In contrast, Table 3 outlines the data collected during testing with the BioSpec Mini-BeadBeater-96, known for its multidirectional homogenization.
|
96-Well Lysis Rack (Benchmark BeadBlaster™ 96 Ball Mill Homogenizer) |
Leakage? |
Cross-Contamination? |
Time Until Failure |
|---|---|---|---|
|
Qiagen PowerBead Pro Plate |
Yes |
Yes |
≤ 10 Minutes |
|
MP Bio Lysing Matrix B, 96-tube Rack |
Yes |
Yes |
≤ 5 Minutes |
|
Omni 96 Deep Well Plate |
Yes |
No |
≤ 10 Minutes |
|
Applied BiosystemsTM MagMAXTM Microbiome Bead Plate |
Yes |
Yes |
≤ 2 Minutes |
|
BioSpec 2ml Deep Well Microplate |
Yes |
Yes |
≤ 5 Minutes |
|
ZR-96 BashingBeadTM Lysis Rack (Barcoded) |
No |
No |
No Failure |
|
ZymoBIOMICS BashingBeadTM Lysis Rack (0.5mm & 0.1mm) |
No |
No |
No Failure |
|
96-Well Lysis Rack (BioSpec Mini-BeadBeater-96) |
Leakage? |
Cross-Contamination? |
Time Until Failure |
|---|---|---|---|
|
Qiagen PowerBead Pro Plate |
Yes |
Yes |
≤ 5 Minutes |
|
MP Bio Lysing Matrix B, 96-tube Rack |
Yes |
Yes |
≤ 5 Minutes |
|
Omni 96 Deep Well Plate |
Yes |
Yes |
≤ 10 Minutes |
|
Applied BiosystemsTM MagMAXTM Microbiome Bead Plate |
Yes |
Yes |
≤ 2 Minutes |
|
BioSpec 2ml Deep Well Microplate |
Yes |
Yes |
≤ 5 Minutes |
|
ZR-96 BashingBeadTM Lysis Rack (Barcoded) |
No |
No |
No Failure |
|
ZymoBIOMICS BashingBeadTM Lysis Rack (0.5mm & 0.1mm) |
No |
No |
No Failure |
Most 96-well lysis racks demonstrated an increase in well-to-well cross-contamination as the shaking speed and motion pattern transitioned from the Benchmark BeadBlasterTM 96 Ball Mill Homogenizer to the BioSpec Mini-BeadBeater-96. Both the ZR-96 BashingBeadTM Lysis Rack (Barcoded) and ZymoBIOMICS BashingBeadTM Lysis Rack (0.5mm & 0.1mm) remained free of any leakage or cross-contamination even after 40 minutes of lysis at 1800 and 2400 RPM, whereas no other 96-well lysis racks withstood more than 10 minutes of lysis without sample leakage or cross-contamination.
Sealing Method Affects Leakage and Cross-Contamination
In this study, the sealing method of the plate had a significant impact on leakage and cross-contamination in the 96-well lysis rack. Our findings indicate a direct relationship between the 96-well lysis rack itself and the sealing method. The stronger the sealing connection, the less likely that a sample well leaks or causes well-to-well cross-contamination in an adjacent well (see supplementary images). Press fit closures and adhesives, like compression mats or PCR-grade seals, that are easily removable show leakage and cross-contamination when subject to low speed mechanical lysis machines. While press fit and adhesive closures for 96-well lysis racks are easy to use and don't require additional equipment, their ease of use does not necessarily ensure effective prevention of leakage or well-to-well cross-contamination. Additionally, the shaking speed, often represented as revolutions per minute (RPM), and processing time also affect the leakage and cross-contamination of a 96-well lysis rack, but were found to be less crucial than the sealing method.
Zymo Research has developed two sealing methods that can withstand both low and high-speed lysis machines, and extended lysis times. The ZymoBIOMICS BashingBeadTM Lysis Rack (0.5mm & 0.1mm) uses a heat sealer and a foil seal to close the 96-well lysis rack. Uniformly applying heat to foil across the 96-well rack allows the foil seal to conform to the shape of the wells and create a secure closure, and ensure each well is sealed consistently. The ZR-96 BashingBeadTM Lysis Rack (Barcoded) uses individual tubes and individual screw caps in a 96-well rack to close each tube. The use of a threaded screw creates a tight seal and are more reliable than alternatives like PCR grade seals. The ZymoBIOMICS BashingBeadTM Lysis Rack (0.5mm & 0.1mm) and ZR-96 BashingBeadTM Lysis Rack (Barcoded) not only prevent well-to-well cross-contamination and sample leakage from individual wells but also establish a secure connection to maintain sample integrity.
The Premier Choice for Prevention of Leakage and Cross-Contamination
Cross-contamination issues have troubled high-profile studies, particularly in the fields of novel virus discovery, molecular anthropology, and clinical diagnostics.6,7 Despite being recognized by many researchers and investigators, cross-contamination remains an understudied and often underreported problem.8 While no specific guidelines exist regarding contamination in studies, some researchers have attempted to identify contaminant profiles using negative controls and then subsequently eliminate these contaminants from their datasets.9 However, this approach, while effective in removing contaminant profiles, carries the risk of eliminating the most significant "real" taxa within the dataset.3 Unfortunately, this can introduce biases and has the potential for misdiagnosing diseases in clinical applications and mischaracterizing samples with low biomass. In low biomass samples, contaminant signals can often dominate the intrinsic signals, especially in samples obtained from sources like the human microbiome.10,11
To tackle these issues with contamination, the utilization of the two Zymo Research Lysis Racks offers an effective solution. Our 96-well lysis racks prevent leakage and cross-contamination during large-scale mechanical lysis, ensuring that researchers can maintain sample integrity throughout all microbial lysis steps. The applications of the ZR-96 BashingBeadTM Lysis Rack (Barcoded) and ZymoBIOMICS BashingBeadTM Lysis Rack (0.5mm & 0.1mm) extend widely across clinical, diagnostic, and research laboratories, making them a valuable tool. For more detailed information about these lysis racks, you can access additional resources via the provided link below.
More About Leakage and Cross-Contamination in 96-Well Lysis Racks
Citations
- Minich JJ, Humphrey G, Benitez RAS, Sanders J, Swafford A, Allen EE, Knight R. 2018. High-throughput miniaturized 16S rRNA amplicon library preparation reduces costs while preserving microbiome integrity. mSystems 3:e00166-18. https://doi.org/10.1128/mSystems.00166-18
- Gu, Wei, Steve Miller, and Charles Y. Chiu. Clinical metagenomic next-generation sequencing for pathogen detection. Annual Review of Pathology: Mechanisms of Disease 14 (2019): 319-338. https://doi.org/10.1146/annurev-pathmechdis-012418-012751
- Minich JJ, Sanders JG, Amir A, Humphrey G, Gilbert JA, Knight R. 2019. Quantifying and understanding well-to-well contamination in microbiome research. mSystems 4:e00186-19. https://doi.org/10.1128/mSystems.00186-19
- Eisenhofer, R., Minich, J. J., Marotz, C., Cooper, A., Knight, R., & Weyrich, L. S. (2019). Contamination in Low Microbial Biomass Microbiome Studies: Issues and Recommendations. Trends in microbiology, 27(2), 105–117. https://doi.org/10.1016/j.tim.2018.11.003
- Lim, M. Y., Song, E. J., Kim, S. H., Lee, J., & Nam, Y. D. (2018). Comparison of DNA extraction methods for human gut microbial community profiling. Systematic and applied microbiology, 41(2), 151–157. https://doi.org/10.1016/j.syapm.2017.11.008
- Salter, S.J., Cox, M.J., Turek, E.M. et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12, 87 (2014). https://doi.org/10.1186/s12915-014-0087-z
- Strong MJ, Xu G, Morici L, Splinter Bon-Durant S, Baddoo M, et al. (2014) Microbial Contamination in Next Generation Sequencing: Implications for Sequence-Based Analysis of Clinical Samples. PLOS Pathogens 10(11): e1004437. https://doi.org/10.1371/journal.ppat.1004437
- Walker AW. 2019. A lot on your plate? Well-to-well contamination as an additional confounder in microbiome sequence analyses. mSystems 4:e00362-19. https://doi.org/10.1128/msystems.00362-19
- Jervis-Bardy, J., Leong, L.E.X., Marri, S. et al. Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data. Microbiome 3, 19 (2015). https://doi.org/10.1186/s40168-015-0083-8
- Nearing, J.T., Comeau, A.M. & Langille, M.G.I. Identifying biases and their potential solutions in human microbiome studies. Microbiome 9, 113 (2021). https://doi.org/10.1186/s40168-021-01059-0
- Weyrich, LS, Farrer, AG, Eisenhofer, R, et al. Laboratory contamination over time during low-biomass sample analysis. Mol Ecol Resour. 2019; 19: 982–996. https://doi.org/10.1111/1755-0998.13011
