Successfully Added to Cart
Customers also bought...
-
 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
 DNA/RNA Shield SafeCollect Swab Collection Kit, 1ml (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA Shield completely inactivates harmful pathogens...
DNA/RNA Shield SafeCollect Swab Collection Kit, 1ml (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA Shield completely inactivates harmful pathogens...
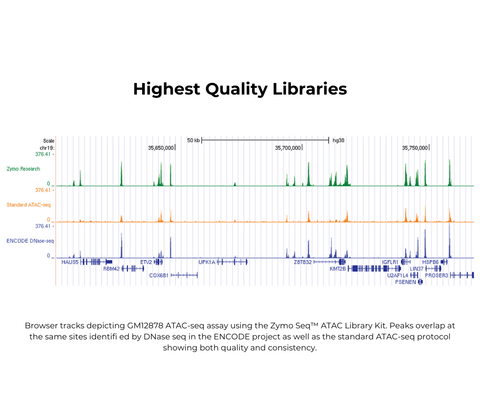
Zymo-Seq ATAC Library Kit
| Cat # | Name | Size | Price | Quantity |
|---|
Highlights
- Ready to Use: Preassembled buffers allow for lightning-fast library preparation in as little as 4 hours without compromising quality.
- Improved Performance: Prepare libraries with 7x less mitochondrial contamination, saving reads and increasing sequencing depth.
- Outstanding Consistency: Produce highly correlated replicates from both fresh and frozen samples.
Documents
Product Description
Technical Specifications
| DNA Input | For optimal results, use 50,000 cells per technical replicate. Cells should present viability (measure of the proportion of live cells within a population) >80%. Recommend using cell counter to measure viability accurately. |
|---|---|
| Equipment Required | Microcentrifuge, Biosafety cabinet, Cell counter, water bath, thermocycler, and thermoshaker with capacity to heat to 37°C and shake at 1000 rpm (We recommend USA Scientific Mixer HC™ with a thermo block that can fit 1.5 mL tubes and 2 mL Low Binding tubes). |
| Library Storage | Libraries eluted in DNA Elution Buffer may be stored at 4°C overnight or at -20°C for long term storage. |
| Processing Time | ~ 4 hours |
| Reagents Not Provided | Trypan blue, PBS, DNase I, cell media, trypsin, FBS, and DMSO. |
| Sequencing Platform Compatibility | Libraries are compatible with Illumina sequencing platforms (HiSeq™, NextSeq™, NovaSeq™). |
Resources
Q1: Can my input be lower than 50,000 live cells?
It is possible to generate libraries with inputs lower than 50k, however, the quality of such libraries is not guaranteed. Typically, we see more PCR duplicates in sequencing data as the amount of input decreases.
Q2: Can my input be higher than 50,000 live cells?
It is possible to generate libraries with inputs higher than 50k, however, the quality of such libraries is not guaranteed.
Q3: Are the Zymo-Seq UDI Primers included individual primers or pre-mixed primer sets?
Each tube contains a premixed forward and reverse primer set that contain a unique i5 and i7 index, respectively. The concentrations of each UDI primer premix is 5µM total (2.5µM each primer).
Q4: Can tissue input be used instead of live cells?
Yes, however, the protocol is optimized for live mammalian cell input and adjustments will have to be made. Please contact (949)-679-1190 ext. 3 or tech@zymoresearch.com for technical support regarding your sample type.
Q5: Why can’t I see my cell/nuclei pellet?
Due to the nature of the assay being very low input, cell and nuclei pellets often will not be visible after they have been spun down. As seen in Section 1 Step 9 of the protocol, if the Low Binding Tubes are placed in the microcentrifuge with the hinge of the tube pointing to the center of the rotor, then the pellet should be found on the side of the tube opposite to the hinge. This location should not change, even when the cell/nuclei pellet is too small to be seen.
| Cat # | Name | Size | Price | |
|---|---|---|---|---|
| D4003T | DNA Clean & Concentrator-5 (Uncapped) | 10 Preps | $31.50 | |
Need help? Contact Us