Successfully Added to Cart
Customers also bought...
-
 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
 DNA/RNA Shield SafeCollect Swab Collection Kit, 1ml (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA Shield completely inactivates harmful pathogens...
DNA/RNA Shield SafeCollect Swab Collection Kit, 1ml (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA Shield completely inactivates harmful pathogens...
Quick-16S Plus NGS Library Prep Kit (V1-V2)
| Cat # | Name | Size | Price | Quantity |
|---|
Highlights
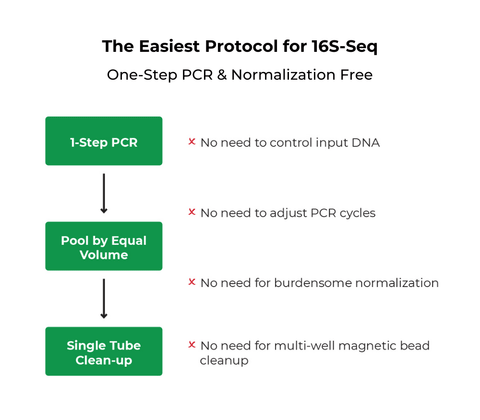
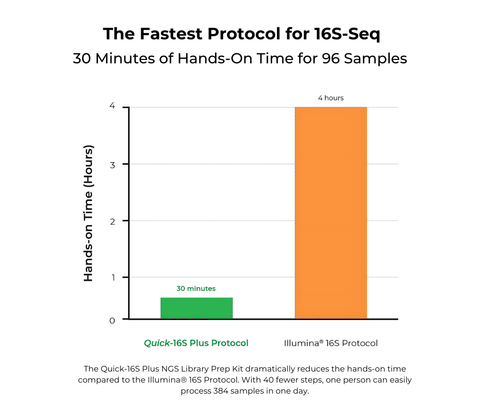
- The most streamlined NGS kit with only 30 minutes of hands-on time for 96 samples.
- 100% automation ready with only a single PCR step and without the need for normalization.
- Real-time PCR enables absolute microbial copy number quantification.
Documents
Product Description
Technical Specifications
| 16s V1-V2 Primer Sequences (adapters not included) | 27f (AGRGTTYGATYMTGGCTCAG, 20 bp) and 341r (CTGCWGCCHCCCGTAGG, 17 bp). |
|---|---|
| Amplicon Size | The final amplicon size after 1-Step PCR (targeted amplification and barcode addition) is ~492 bp. |
| Barcode Sequences | 10 bp barcodes are available to download here or by visiting the Documentation section of this Product Page. |
| Index Primers | Unique dual index (barcodes) to label individual samples. |
| Required Equipment | Microcentrifuge, plate spinner (centrifuge), 96-well real-time quantitative PCR system (SYBR Green compatible) or standard PCR system, and 96-well real-time PCR plates. |
| Sample Input | Purified microbial DNA ≤100 ng, free of PCR inhibitors. |
| Sequencing Platform | Illumina MiSeq® or NextSeq 1000/2000® without the need to add custom sequencing primers. We recommend the MiSeq® Reagent Kit v3 (600-cycle) or NextSeq 1000/2000® Reagent Kit P1/P2 (600-cycle). For assistance with sample sheet setup, see Appendix F. |
Resources
Q1: What samples are compatible with this kit?
Purified total DNA from any organism free from PCR inhibitors. This system has been used by Zymo Research’s Microbiomics Services team to process samples from human, water, soil, food, plants, feces, biofilms, and much more.
Q2: What is the minimum DNA input for this kit?
The system has been tested with inputs as low as 100 femtograms with no significant impact to the performance of the kit.
Q3: Why does the kit use only a single PCR amplification step?
This single PCR step combines targeted amplification of the microbial genome and the barcoding index PCR, which allows for a simple and fast workflow.
Q4: How does the kit achieve normalization?
The combination of using Equalase qPCR Premix to yield roughly equal amounts of libraries and a carefully curated barcode index list results in similar raw sequencing reads across samples.
Q5: What is the difference between the two standards included in the kit?
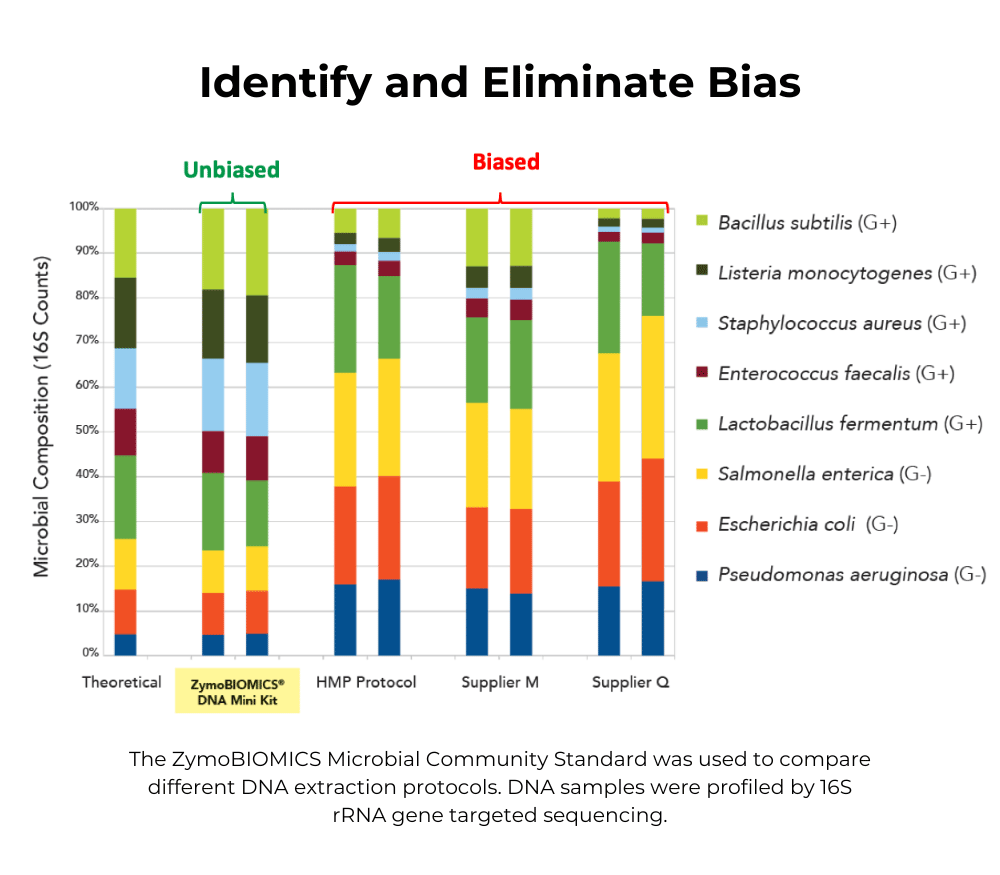
The ZymoBIOMICS™ Microbial Community DNA Standard consists of purified DNA from 8 bacteria and 2 yeasts and is useful for assessing community profile bias. The ZymoBIOMICS™ 16S/ITS qPCR Standard consists of a plasmid that has a single bacterial and single fungal target and is useful for quantifying copy number. Both are useful as positive controls for the library prep process.
Q6: Why is the library amplicon size not well defined in DNA fragment analysis?
Equalase amplification and the library prep design may cause some library products to not anneal well, causing a lack of a tight band. These libraries are perfectly fine because preparation for Illumina sequencing includes denaturing libraries into single strands.
Q7: What samples are compatible with this kit?
Purified total DNA from any organism free from PCR inhibitors. This system has been used by Zymo Research’s Microbiomics Services team to process samples from human, water, soil, food, plants, feces, biofilms, and much more.
Q8: What is the minimum DNA input for this kit?
The system has been tested with inputs as low as 100 femtograms with no significant impact to the performance of the kit.
Q9: Why does the kit use only a single PCR amplification step?
This single PCR step combines targeted amplification of the microbial genome and the barcoding index PCR, which allows for a simple and fast workflow.
Q10: How does the kit achieve normalization?
The combination of using Equalase qPCR Premix to yield roughly equal amounts of libraries and a carefully curated barcode index list results in similar raw sequencing reads across samples.
Q11: What is the difference between the two standards included in the kit?
The ZymoBIOMICS™ Microbial Community DNA Standard consists of purified DNA from 8 bacteria and 2 yeasts and is useful for assessing community profile bias. The ZymoBIOMICS™ 16S/ITS qPCR Standard consists of a plasmid that has a single bacterial and single fungal target and is useful for quantifying copy number. Both are useful as positive controls for the library prep process.
Q12: Why is the library amplicon size not well defined in DNA fragment analysis?
Equalase amplification and the library prep design may cause some library products to not anneal well, causing a lack of a tight band. These libraries are perfectly fine because preparation for Illumina sequencing includes denaturing libraries into single strands.
| Cat # | Name | Size | Price | |
|---|---|---|---|---|
| D4302-5-10 | ZymoBIOMICS DNase/RNase Free Water | 10 ml | $32.70 | |
| D6305 | ZymoBIOMICS Microbial Community DNA Standard | 200ng | $121.30 | |
| D6306 | ZymoBIOMICS Microbial Community DNA Standard | 2000ng | $242.80 | |
Related Products
D6300
ZymoBIOMICS Microbial Community Standard
Microbial composition profiling techniques powered by Next-Generation sequencing are becoming routine in microbiomics and metagenomics studies. However, thes...
D4300, D4300T, D4304
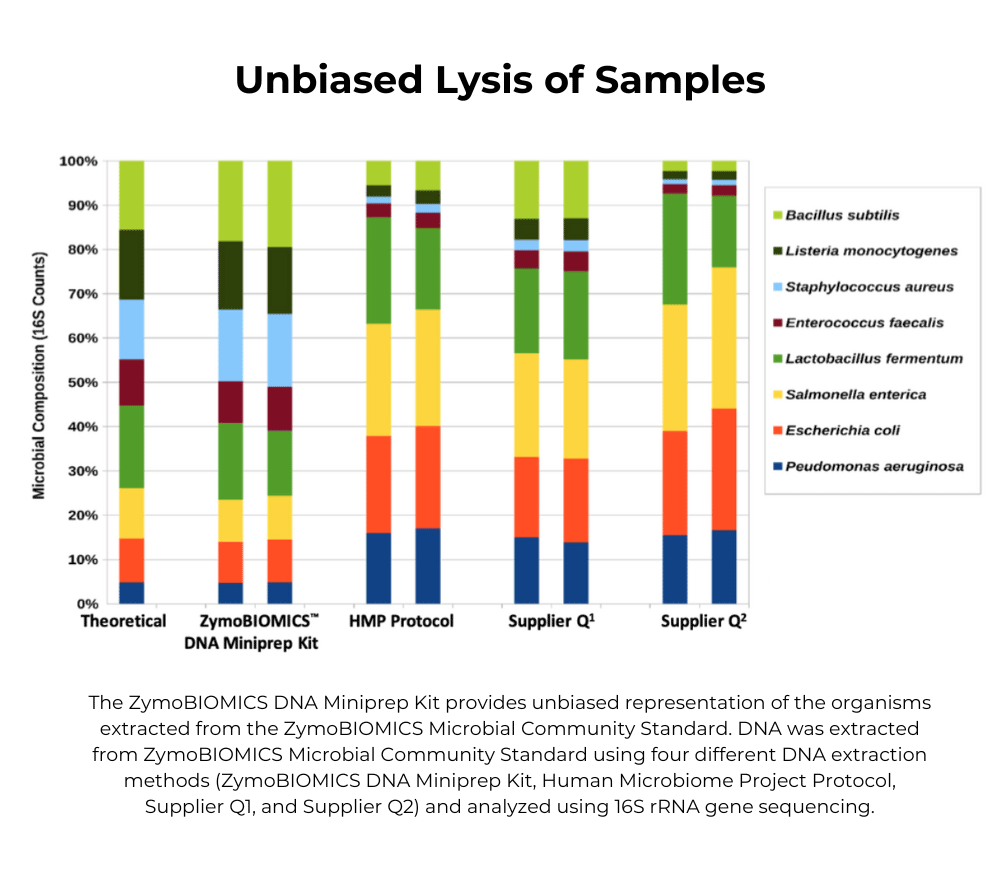
ZymoBIOMICS DNA Miniprep Kit
Efficient microbial DNA purification from low sample inputs for highly accurate microbiome and metagenome analyses.

R1160, R1161, R1160-E...
DNA/RNA Shield SafeCollect Swab Collection Kit
DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly, spill-free device allowing for safe, convenient collection of swab specimen samples. Ideal...
Need help? Contact Us