Low-input RRBS DNA Methylation Profiling
From as Little as 10ng of DNA
Reduced Representation Bisulfite Sequencing (RRBS) Is Informative and Efficient
Needs only ~ 10% of sequencing reads compared to whole-genome bisulfite sequencing
Covers ≥70% of promoters, CpG islands, gene bodies, and ~ 35% of enhancers*
Ideal for high throughput DNA methylation screening at single nucleotide resolution
RRBS Is Applicable Across Diverse Species

RRBS Has Many Popular Applications
Developmental Biology

Cancer Research
Drug Response Study

Biomarker Discovery

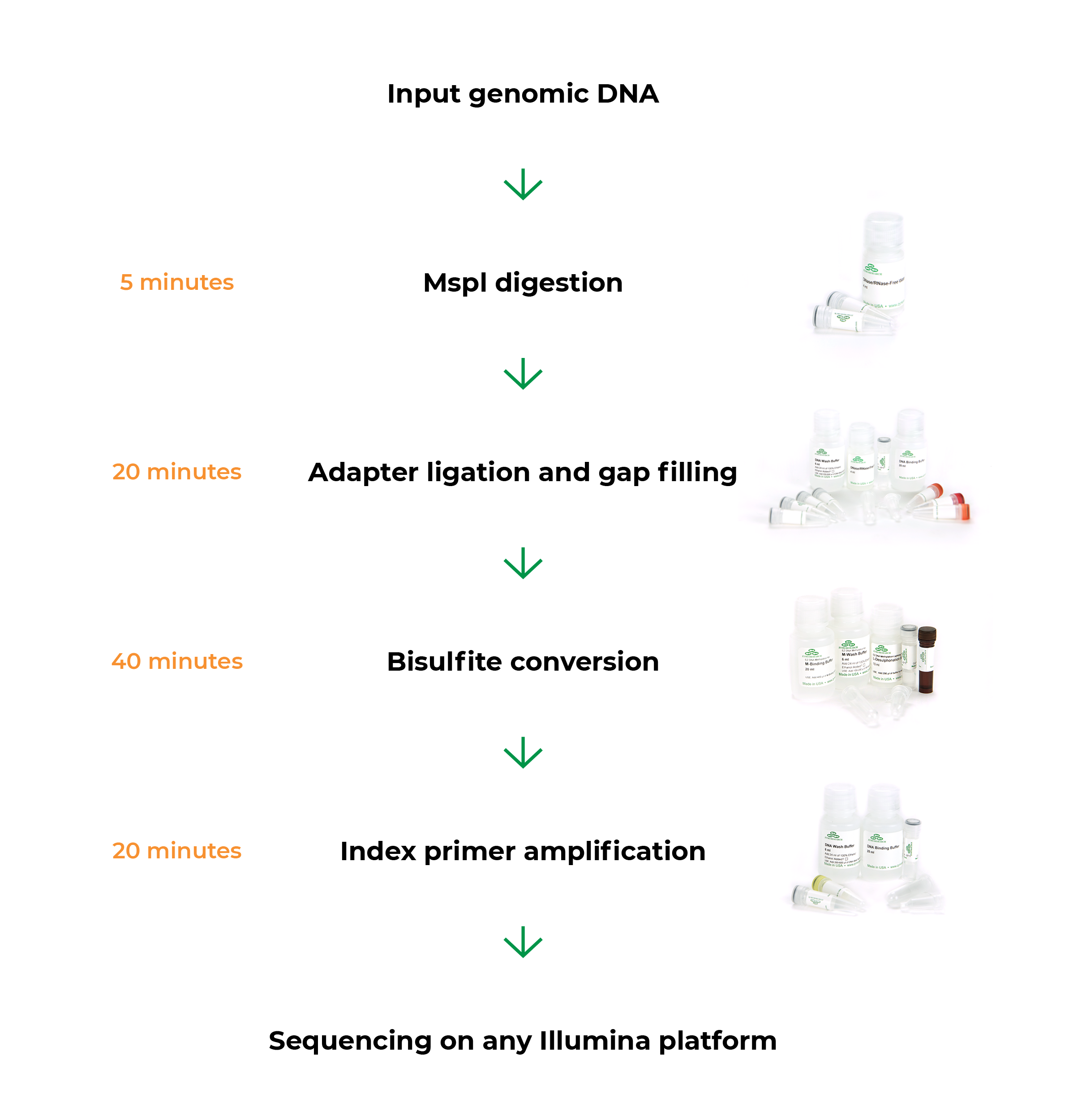
Zymo-Seq RRBS Provides A Simple Solution for NGS Library Preparation
The workflow minimizes hands-on time.
Zymo-Seq RRBS Detects a Significant Number of CpG Sites

Distribution of CpG sites detected (≥ 5X, green dots) in a library constructed with Zymo-Seq RRBS Library Kit across the human chromosomes using 10 ng of human liver genomic DNA
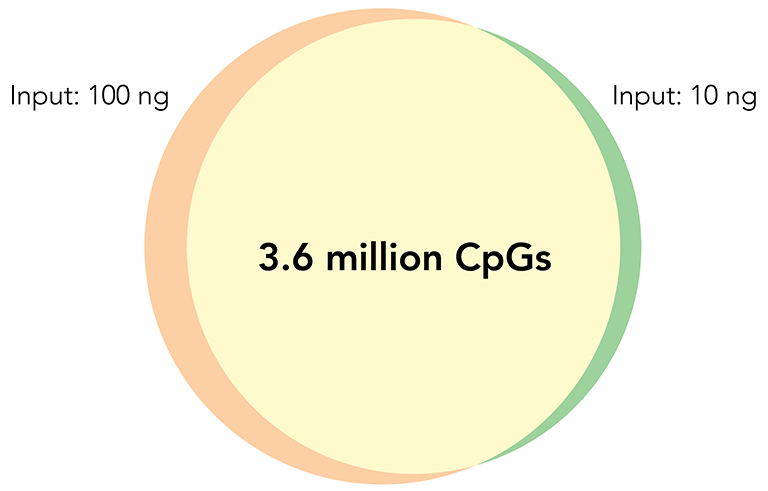
Zymo-Seq RRBS Delivers Reproducible Results Regardless of Input Amount
CpG Sites Overlap
Methylation Ratio Correlation
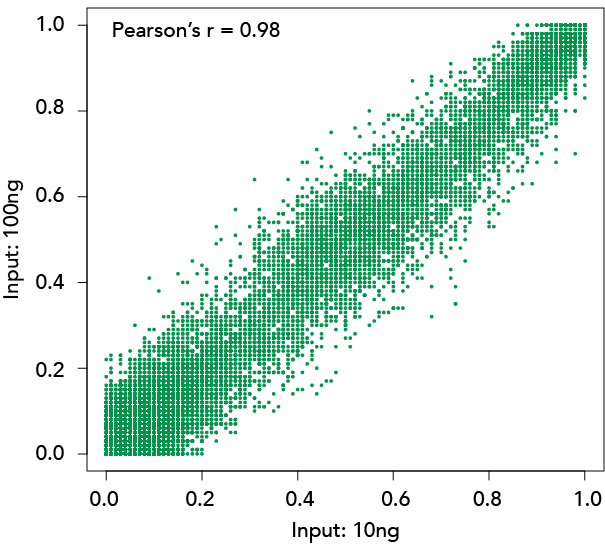
Libraries constructed with Zymo-Seq RRBS Library Kit using 10 ng and 100 ng of human liver genomic DNA.
Left: Overlap of CpG sites detected (≥5X). Right: Methylation ratio correlation of CpG sites detected (≥50X)
Simplify Your Research With The Zymo-Seq RRBS Library Kit
* Enhancers derived from H3K4me2 peaks. Data based on libraries generated using Zymo-Seq RRBS Library Kit with various amount of human liver genomic DNA.