Metatranscriptomics Sequencing Service
Reveal Microbial Dynamics with Gene Expression Profiling
Universal rRNA Depletion
Deplete host and bacterial ribosomal RNA (rRNA) from any sample type

Extreme Flexibility
Comprehensive gene expression analysis

Microbiome Master
Detect RNA from any kingdom (Bacteria, Archaea, Eukarya, and viruses)
High quality RNA from any sample type using unbiased mechanical lysis
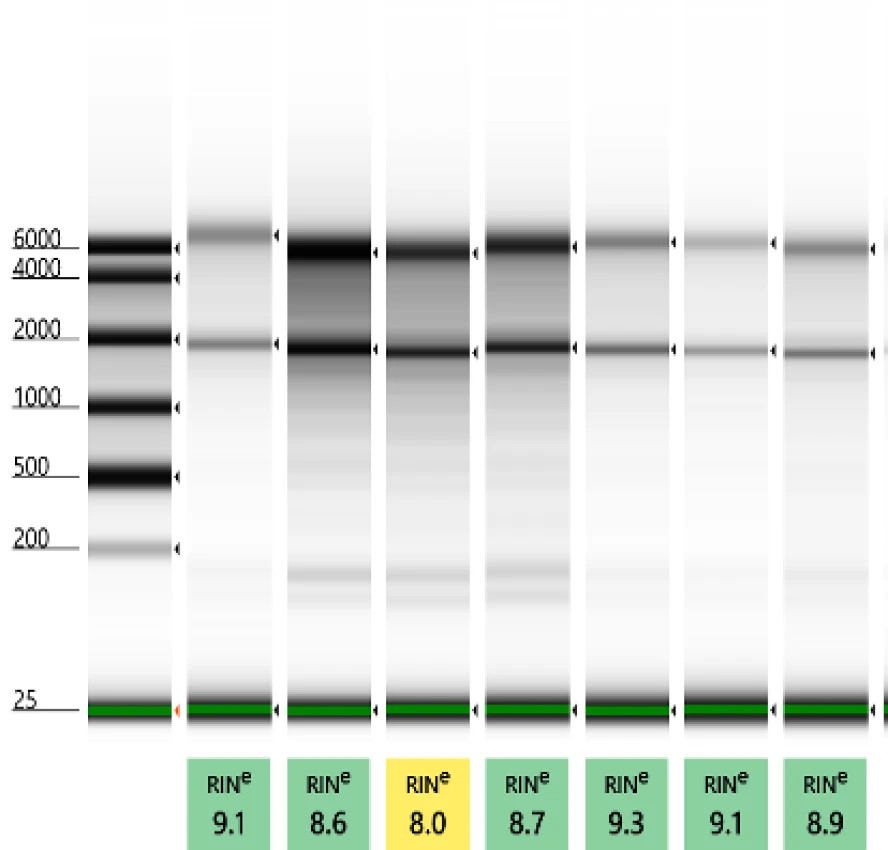
Fragment size distribution and RIN score of RNA extracted using ZymoBIOMICS MagBead RNA Kit (R2137) or ZymoBIOMICS RNA Miniprep Kit (R2001).
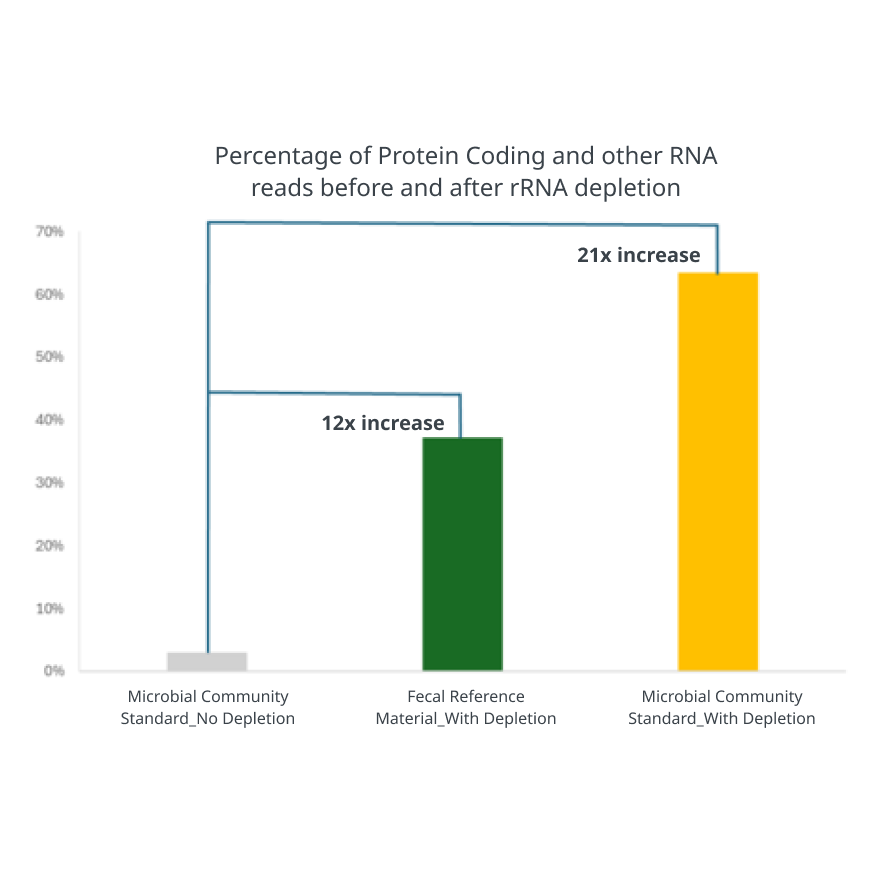
More relevant data with universal rRNA depletion
Ribosomal RNA depletion and RNA library prep was performed on ZymoBIOMICS Community Standard (D6300) and Fecal Reference Material (D6323) using Zymo-Seq RiboFree Total RNA Library Kit (R3000).
Metabolic Pathway and Gene Expression Analysis
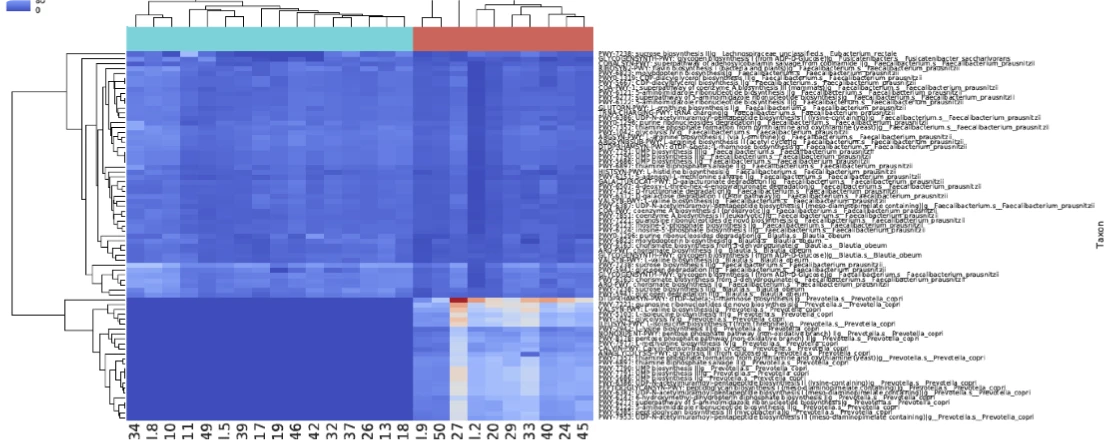
Multi-Kingdom Taxonomy Identification
Reveal Microbial Dynamics with Gene Expression Profiling
Bacteria
Fungi
Viruses

Protists
The Zymo Research database contains over 77,000 reference strains across all kingdoms (Bacteria, Archaea, Eukaryote and DNA Viruses). Proprietary algorithm eliminates the risks of false positives in taxonomy assignment.
Metatranscriptomics Sequencing Made Simple
How our Straightforward, User-Friendly Service Works

Inquire Below
Send Us Your Samples

Receive A User-Friendly
Analysis Report
Trusted By Leading Institutions
We are honored to have collaborated and provided next-generation analysis services with the most prestigious institutions, agencies and companies in the world.
Technical Specifications
| Sequencing Platform | Illumina NovaSeq X™ 10B or 25B Reagent kit (2x150bp) |
|---|---|
| Sample Collection |
Raw Sample Various sample collection devices provided upon request RNA sample >10 ng/µl (by Fluorescence RNA quantification) with RIN score >7 Please ensure it is PCR inhibitor-free. |
| RNA Extraction |
ZymoBIOMICS 96 Magbead RNA (R2137)
ZymoBIOMICS RNA Miniprep (R2001)
|
| Library Prep |
Zymo-Seq RiboFree Total RNA Library Kit (R3003)
Unique Dual Index (UDI)
|
| Quality Control | A positive control for RNA extraction (ZymoBIOMICS Microbial Community Standard, D6300) is included in each project as a quality control, without additional charge. Negative extraction controls and negative PCR controls are monitored and can be sequenced upon request. |
| Sequencing Analysis | Kmer signature-based analysis using Sourmash with antimicrobial resistance using Diamond and functional pathway analysis using Humann3 |
| Taxonomy Resolution |
Bacteria and Archaea: strain-level
Fungi and Viruses: species-level
|
|---|---|
| Reference Database | Full GTDB database (R07-RS207) was used for bacterial and archaeal identification. Pre-formatted GenBank databases (v2022.03) provided by Sourmash were used for virus, protozoa, and fungi identification |
| Data Analysis |
Taxonomy Profiling Taxa abundance bar plot and heatmap Alpha diversity boxplot Various sample collection devices provided upon request |
| Statistical Analysis | LEfSe analysis uncovers taxa/traits that are differentially distributed among defined sample groups. |
| Example Report | Our Sample Report can be viewed here. |
| Data Storage | Raw sequencing data and analysis report are stored for up to 3 months. |
Frequently Asked Questions
Is there a minimum sample number?
There is no sample minimum required in order to submit to our Metatranscriptomics Sequencing Service.
What are the requirements for raw samples?
We recommend sending 200-500 mg for solid samples or 10-15mL of liquid samples in general. This may vary depending on the specific sample type.
What are the requirements for RNA samples?
The minimum volume of DNA required is 50 µl with a recommended concentration of 10-50 ng/µl. If the concentration is lower, we can often still successfully sequence as our pipeline is quite robust.
Which sample types are accepted?
We have processed a wide variety of sample types, from human tissue samples, environmental samples, to food samples. If you have questions or concerns, please feel free to fill out an inquiry or contact us.
How do you control for run-to-run sequencing variation?
Because of our included quality control standard, it is possible to batch samples over time and have confidence that the samples were processed in the same way. We recently completed a project that had a total of >2,000 samples processed over a period of 6 months. By comparing the results from the standard for each group of samples, we had confidence that the results from different batches could be compared together.
Why Partnering with Zymo Research for Microbiome Applications is Your Key to Success.
Metatranscriptomics has emerged as a powerful approach to unravel the functional dynamics of complex microbial communities. By capturing real-time gene expression patterns, metatranscriptomic sequencing offers unprecedented insights into microbial activities and ecological functions. At Zymo Research, we've harnessed this cutting-edge technique to provide a comprehensive metatranscriptomic sequencing service, revolutionizing microbiome analysis.
The metatranscriptomic sequencing workflow begins with sample collection, ensuring the preservation of microbial profiles. Total RNA isolation, rRNA depletion, library preparation, and high-throughput sequencing follow, each step optimized and validated to minimize bias and ensure accurate results. With stringent quality control measures in place, we guarantee the reliability of our data, empowering researchers with trustworthy insights.
Zymo Research's legacy in microbiome research is built on innovative products designed to meet the unique needs of researchers. Our DNA/RNA Shield ensures unbiased sample collection, while our ZymoBIOMICS RNA kits enable unbiased lysis. These products, coupled with our microbiome standards, ensure superior process quality control, laying the foundation for our metatranscriptomic sequencing service.
Central to our metatranscriptomic sequencing service is a comprehensive and user-friendly bioinformatics report. This report includes strain-level taxonomy profiling, metabolic gene family and pathway profiling, virulence gene and antibiotic resistance gene identification, and statistical analysis. Our advanced taxonomy assignment algorithm minimizes false positives, enhancing the accuracy of microbial identification.
The applications of metatranscriptome sequencing are vast and impactful. In microbial ecology research, it aids in characterizing functionally active bacteria and elucidating community metabolic interactions. In clinical research, it uncovers immune responses and autoimmune processes, while also offering insights into innovative drug delivery resources. Additionally, it improves food quality by providing data on microbial metabolic behaviors, paving the way for safer and healthier food products.
Zymo Research's metatranscriptomic sequencing service represents a leap forward in microbiome analysis. With a robust workflow, innovative products, and comprehensive bioinformatics analysis, we empower researchers to unlock the functional mysteries of microbial communities. From environmental studies to clinical applications, metatranscriptomic sequencing holds the key to transformative discoveries, and Zymo Research is at the forefront of this revolution.