
Long-Read Bacterial Whole Genome Assembly
Assembling Complete Genomes from Any Bacterial Culture
High Molecular Weight DNA Extraction
HMW DNA from DNA extraction with mechanical microbial lysis
Comprehensive Assembly Analysis
Sequence assembly, strain-level taxonomy assignment, annotation, etc.

Powered by PacBio HiFi Sequencing
Entrust your samples to Certified PacBio Certified Provider
High Molecular Weight DNA Extraction With Mechanical Lysis
Confidently Obtain High-quality DNA Even From Hard-to-Lyse Bacteria
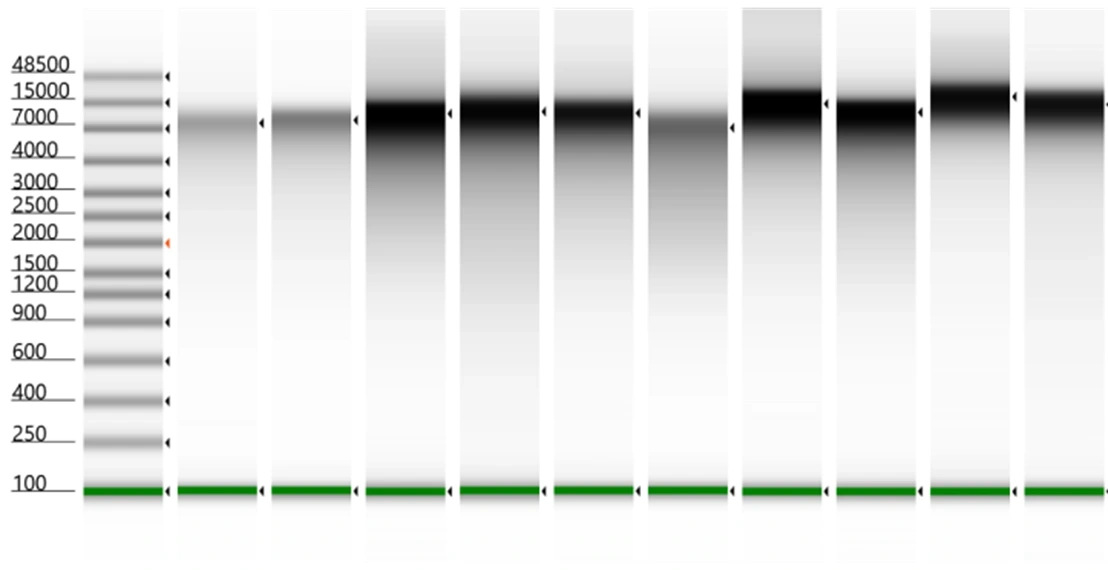
DNA fragment size distribution from bacterial isolates with varying levels of recalcitrance. The samples were processed with the ZymoBIOMICS 96 DNA MagBead Kit and Miniprep Kit to ensure efficient extraction.
Highly Accurate Assemblies with Long-Read HiFi Sequencing
Genome Assemblies are consistently > 99.99% accurate

Organism
Data obtained from HiFi Sequencing has consistent Q Value > 40.
Sophisticated Genome Assembly & Annotation
Publication-ready high-fidelity genome and annotations
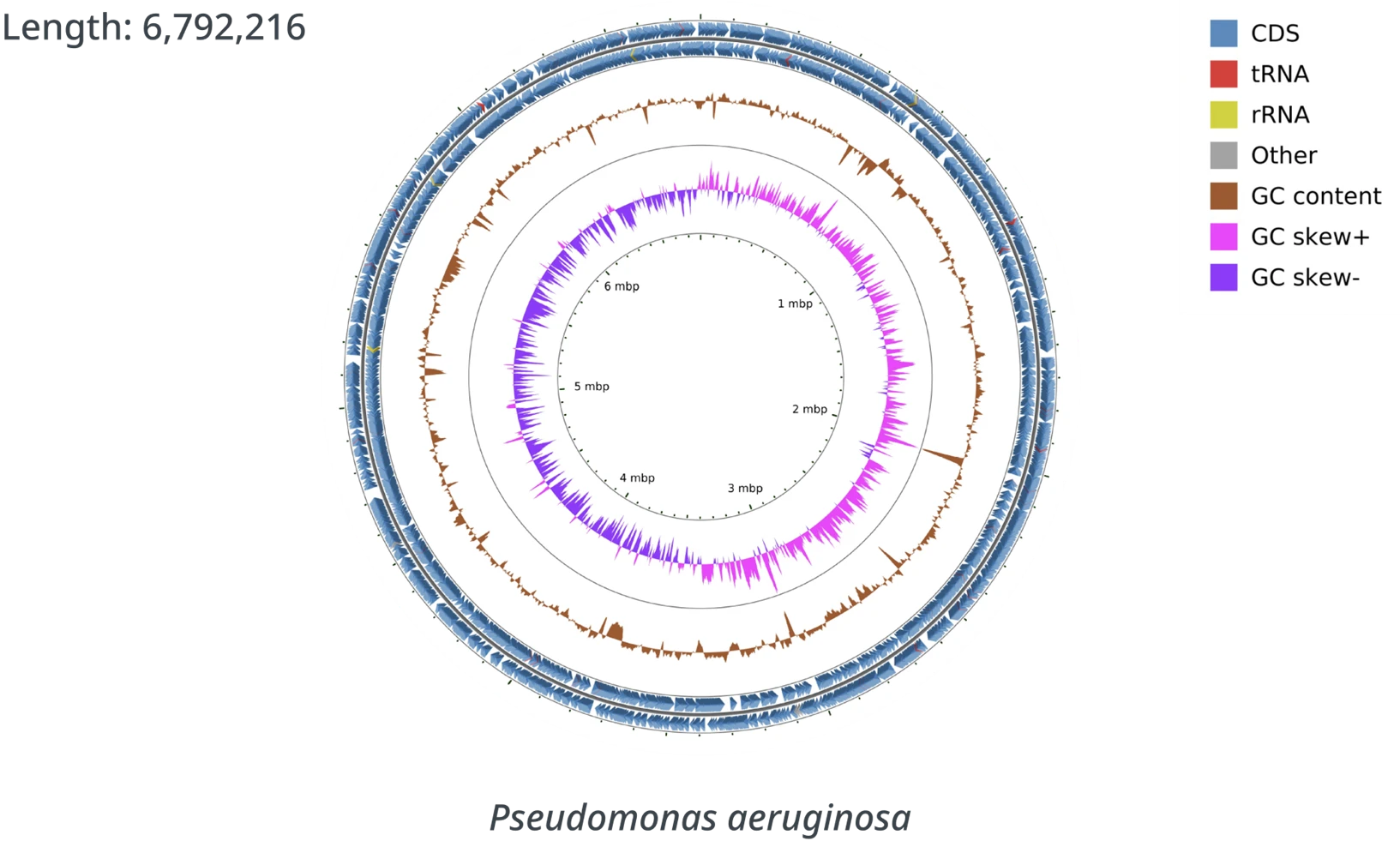
Simplifying Genome Assembly Interpretation

The Genome Assembly report provides sequence read distribution, assembly result summary, taxonomy prediction, genome annotation summary, antimicrobial resistance genes identification, and circular genome map. Our team of expert bioinformaticians are here to help you get the most out of your data.
Click below to view a sample report:
Bacterial Whole Genome Assembly Made Simple
How our Straightforward, User-Friendly Service Works

Inquire Below
Send Us Your Samples

Receive A User-Friendly
Analysis Report
Bacterial Whole Genome Assembly Technical Specifications
| Sequencing Platform | PacBio Sequel IIe or Revio |
|---|---|
| Sequencing Kit | SMRT Cell 8M or 24M (PacBio) |
| Accepted Sample Types | Extracted genomic DNA, or raw samples |
| DNA Extraction |
ZymoBIOMICS Magbead DNA/RNA kit (R2135)
ZymoBIOMICS RNA Miniprep Kit (R2001)
|
| Library Prep |
SMRTbell prep kit 3.0 (with SMRTbell cleanup beads) or HiFi plex prep kit 96 (PacBio)
Barcoded Adapter Plate 3.0 (PacBio)
|
| Sequencing Analysis | PacBio HiFi reads are assembled with flye v2.9.2 and hifiasm v0.19.8. The best assembly was selected and reported. The assembled draft or complete genomes are annotated with prokka v1.14.5. The taxa of the genome is predicted using gtdbtk v2.3.2. Antimicrobial resistance genes and virulence genes are identified by searching reads against databases curated from NCBI using Diamond |
| Data Visualization | Circular genome maps are prepared using CGview |




