ChIP-Seq Analysis Service
Personalized Project Design with Comprehensive Bioinformatic Analysis
ChIP-Seq analysis service is a powerful tool for genome-wide mapping of histone modifications, protein DNA interactions, and identifying consensus protein binding sites in DNA.
Full Chromatin Immunoprecipitation Services
ChIP-Seq Analysis Service Features
- Customized Help With Design Plan
- End-to-End Support From an Expert Team
- In house Next-Gen Sequencing
- Rapid Turnaround Times
- Comprehensive Bioinformatics
- Low Sample Input Compatible
- Customized Help With Design Plan
- In house Next-Gen Sequencing
- Comprehensive Bioinformatics
- End-to-End Support From an Expert Team
- Rapid Turnaround Times
- Low Sample Input Compatible
High Quality Data Generation
Access To Experienced Scientists
Our dedicated, PhD led team has over 20 years of epigenetic expertise. With the Zymo Research ChIP-Seq analysis service, all you need to do is submit your samples and let the Zymo Research service team process them using optimized proprietary enrichment procedures and NGS library sequencing and analysis.
If you have a completed ChIP assay, Zymo Research's qualified scientists can assist you in library construction and Next-Gen Sequencing.
Publication-Ready Data At Your Fingertips
Zymo Research's ChIP-Seq analysis service includes comprehensive in-house bioinformatics and statistical analyses as well as group comparisons, presented in a user-friendly and interactive report.
Let Zymo Research do the bioinformatics so you don't have to!
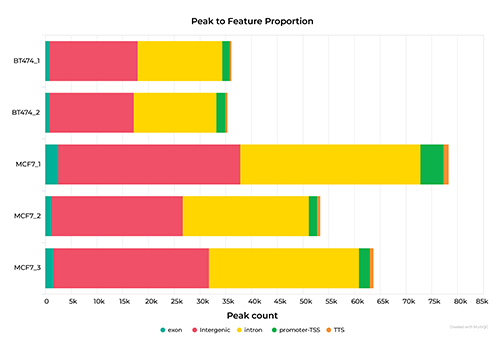
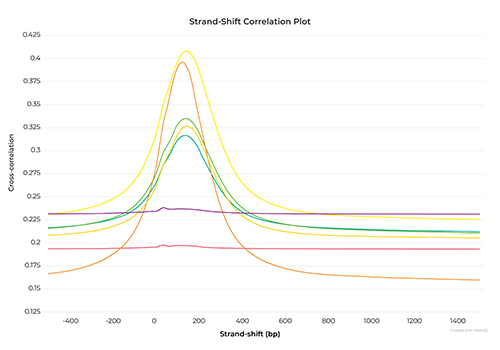
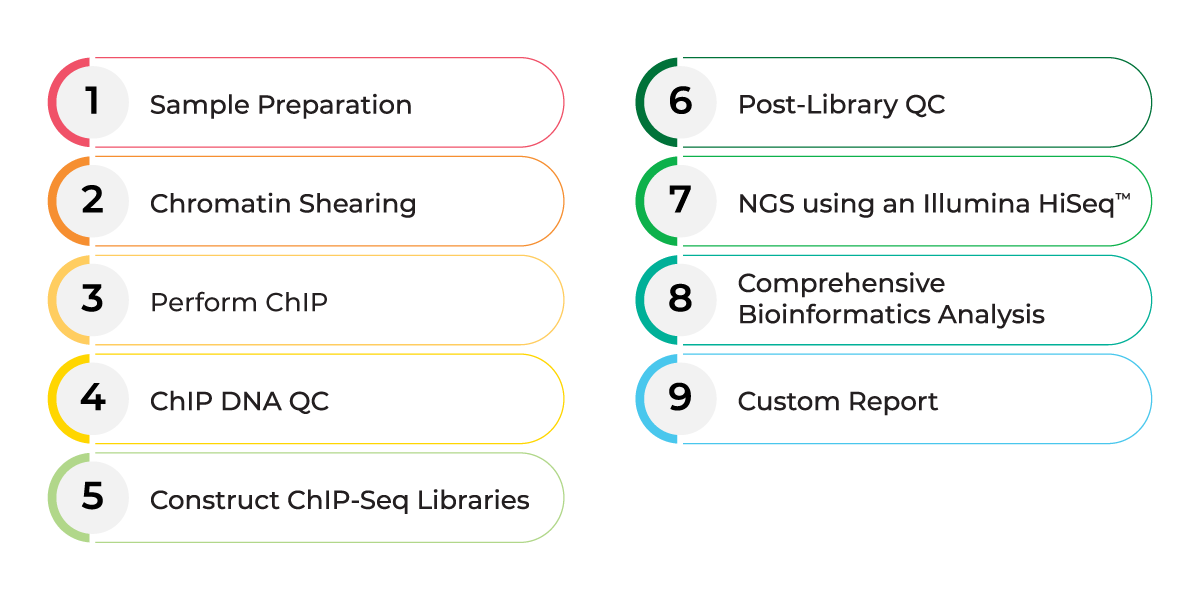
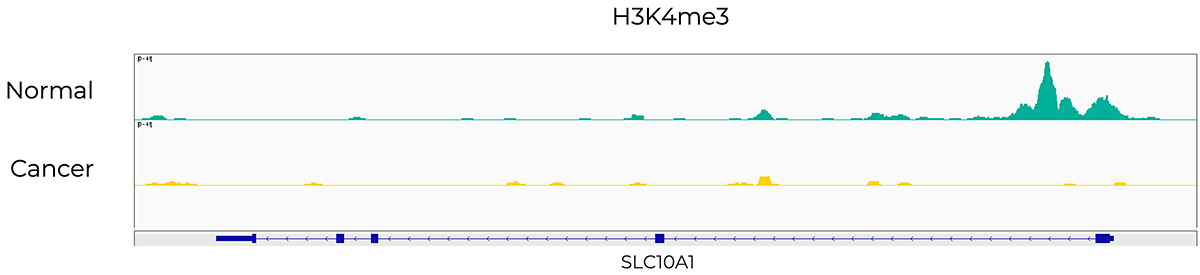
Introduction to ChIP-Seq Data Analysis
Have questions about the ChIP-Seq analysis service process?
Learn more about ChIP-Seq and its analysis below.
Cited in Major Publications
Featured Citations
ChIP-Seq Analysis Service Citations
- Apostolova, G., et al. Satb2 determines miRNA expression and long-term memory in the adult central nervous system. eLife 5, (2016).
- Cattini, P. A., et al. Expression of placental members of the human growth hormone gene family is increased in response to sequential inhibition of DNA methylation and histone deacetylation. BioResearch Open Access 4, 1 (2015).
- Firestein, G. S., et al. Comprehensive epigenetic landscape of rheumatoid arthritis fibroblast-like synoviocytes. Nature Communications 9, 1921 (2018).
- Gilfillan, G. D., et al. A comparative study of ChIP-Seq sequencing library preparation methods. BMC Genomics 17, 816 (2016).
- Grueter, C. E., et al. Disruption of cardiac Med1 inhibits RNA polymerase ll promoter occupancy and promotes chromatin remodeling. American Journal of Physiology- Heart and Circulatory Physiology 316, 2 (2018).
- Martin, R.C., Vining, K. & Dombrowski, J.E. Genome-wide (ChIP-seq) identification of target genes regulated by BdbZIP10 during paraquat-induced oxidative stress. BMC Plant Biol 18, 58 (2018).
- Schulte, J. H., et al. Targeting MYCN-driven transcription by BET-bromodomain inhibition. Clinical Cancer Research 22, 10 (2016).
- Sun, Z., Chadwick, B.P. Loss of SETDB1 decompacts the inactive X chromosome in part through reactivation of an enhancer in the IL1RAPL1 gene. Epigenetics and Chromatin 11, 45 (2018).