Successfully Added to Cart
Customers also bought...
-
 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
 DNA/RNA Shield SafeCollect Swab Collection Kit, 1ml (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA Shield completely inactivates harmful pathogens...
DNA/RNA Shield SafeCollect Swab Collection Kit, 1ml (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA Shield completely inactivates harmful pathogens...
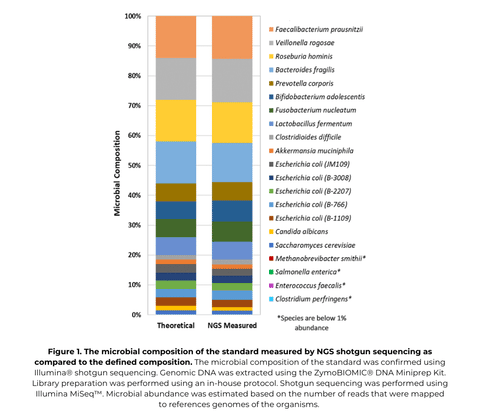
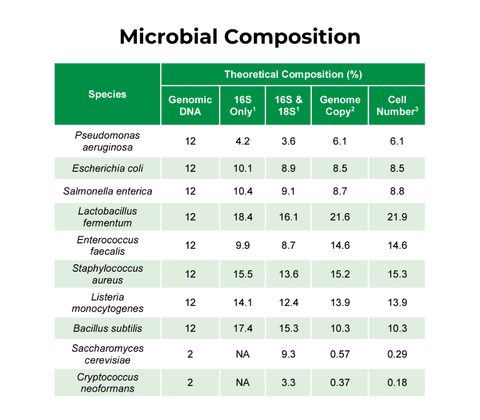
ZymoBIOMICS Gut Microbiome Standard
| Cat # | Name | Size | Price | Quantity |
|---|
Highlights
- True to Life: comprised of 21 different strains to mimic the human gut microbiome.
- Accurate Composition: allows for benchmarking and validation of NGS microbiome workflows.
- Cross Kingdom Representation: includes Bacteria, Fungi, and Archaea.
Documents
Product Description
Technical Specifications
| Biosafety | BSL 1. Microbes have been fully inactivated. |
|---|---|
| Impurity Level | < 0.01% foreign microbial DNA |
| Reference Genomes | 16S&18S rRNA genes https://s3.amazonaws.com/zymo-files/BioPool/D6331.refseq.zip |
| Relative-abundance Deviation | <15% |
| Storage Solution | 2X DNA/RNA Shield (Cat. No. R1200-125). |
| Total Cell Concentration | ~3.94 x 109 cells/ml |
Resources
Q1: Should I use D6300 Microbial Community Standard or D6331 Gut Microbiome Standard?
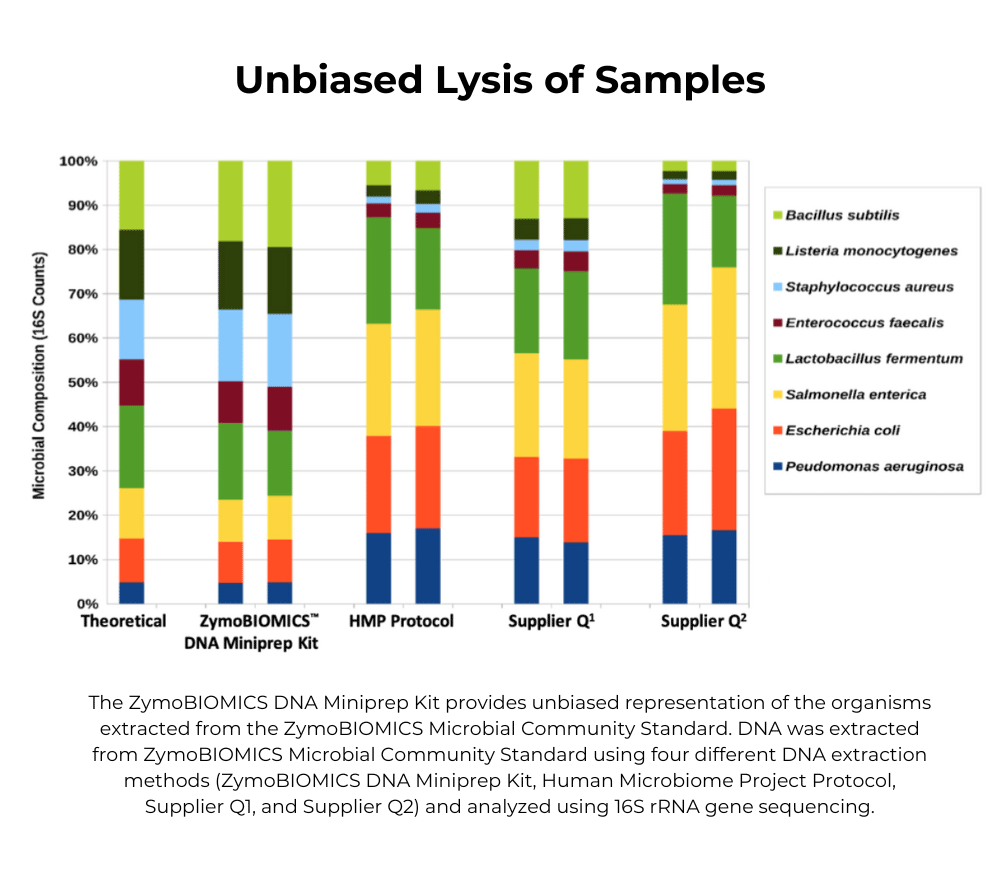
For users that are setting up a new microbiomics workflow, we recommend starting with the D6300 Microbial Community Standard. The lower complexity of the standard will enable the user to more easily detect existing biases in the workflow (i.e. lysis during DNA extraction). Users who have validated using the D6300 already may use the D6331 to then fine-tune their workflows for gut samples.
Related Products

R1133-1, R1133-10, R1138...
Bunny Wipe Fecal Sample Collector
User-friendly at home fecal collection with the Bunny Wipe fecal sample collector and the DNA/RNA Shield Fecal Collection Tube.
D4300, D4300T, D4304
ZymoBIOMICS DNA Miniprep Kit
Efficient microbial DNA purification from low sample inputs for highly accurate microbiome and metagenome analyses.
D6421-PS1, D6421-PS2, D6421-PS3...
Quick-16S Plus NGS Library Prep Kit (V3-V4, UDI)
The Quick-16S Plus NGS Library Prep Kit (V3-V4) is the fastest and simplest NGS library prep kit targeting the 16S rRNA gene for microbiome profile analysis.
Need help? Contact Us