
Total RNA-Seq Library Prep Kit for Any Organism
Zymo-Seq RiboFree Total RNA Library Kit
Comprehensive Gene Detection for Every Transcriptome & Beyond
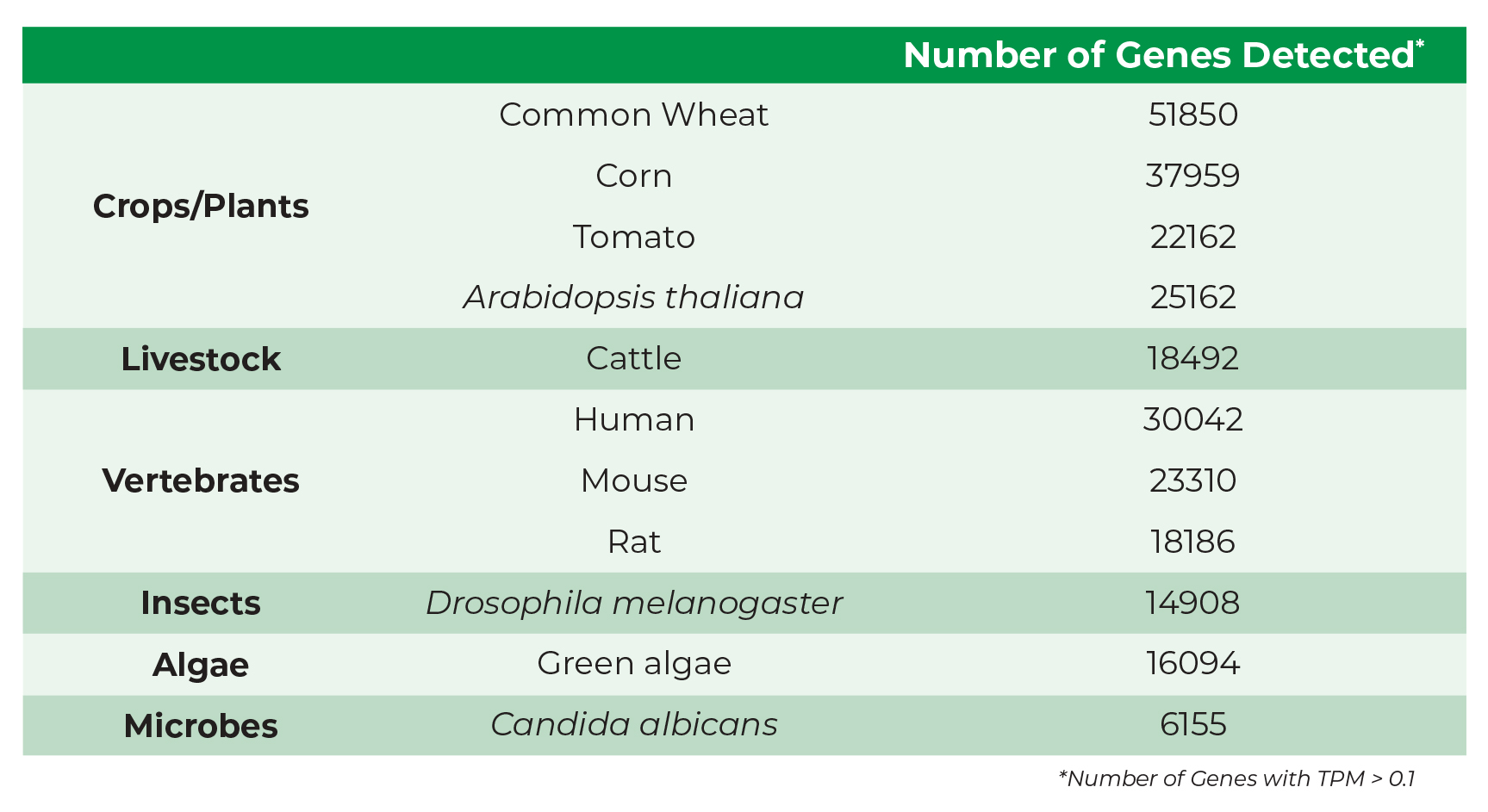
The Zymo-Seq RiboFree Kit Detects an Exceptional Number of Genes. Total RNA extracted from various species were used as inputs for library preparation using a single standard RiboFree protocol. The libraries were paired-end sequenced. STAR-aligned reads were quantified at the gene level using featureCounts.
Maximize Your Sequencing Efficiency
>90% rRNA Depletion Across Organisms
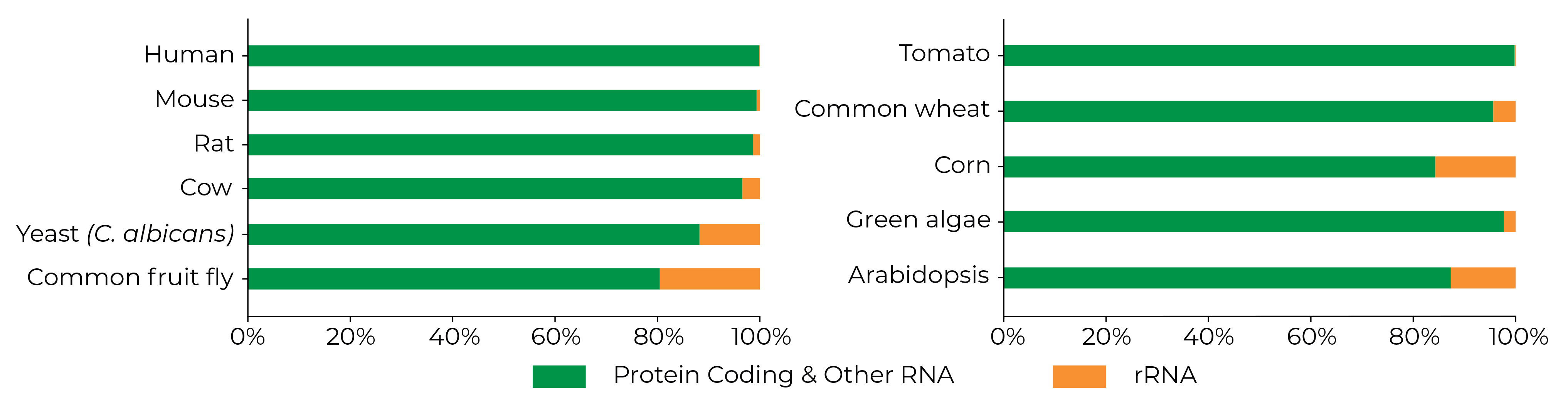
The Zymo-Seq RiboFree Kit Produces Dense Coverage of Protein Coding and Other Transcripts. Classification of the STAR-aligned reads was based on Ensembl annotations and RepeatMasker rRNA tracks from UCSC genome browser when applicable.
Unparalleled Multi-Species Compatibility
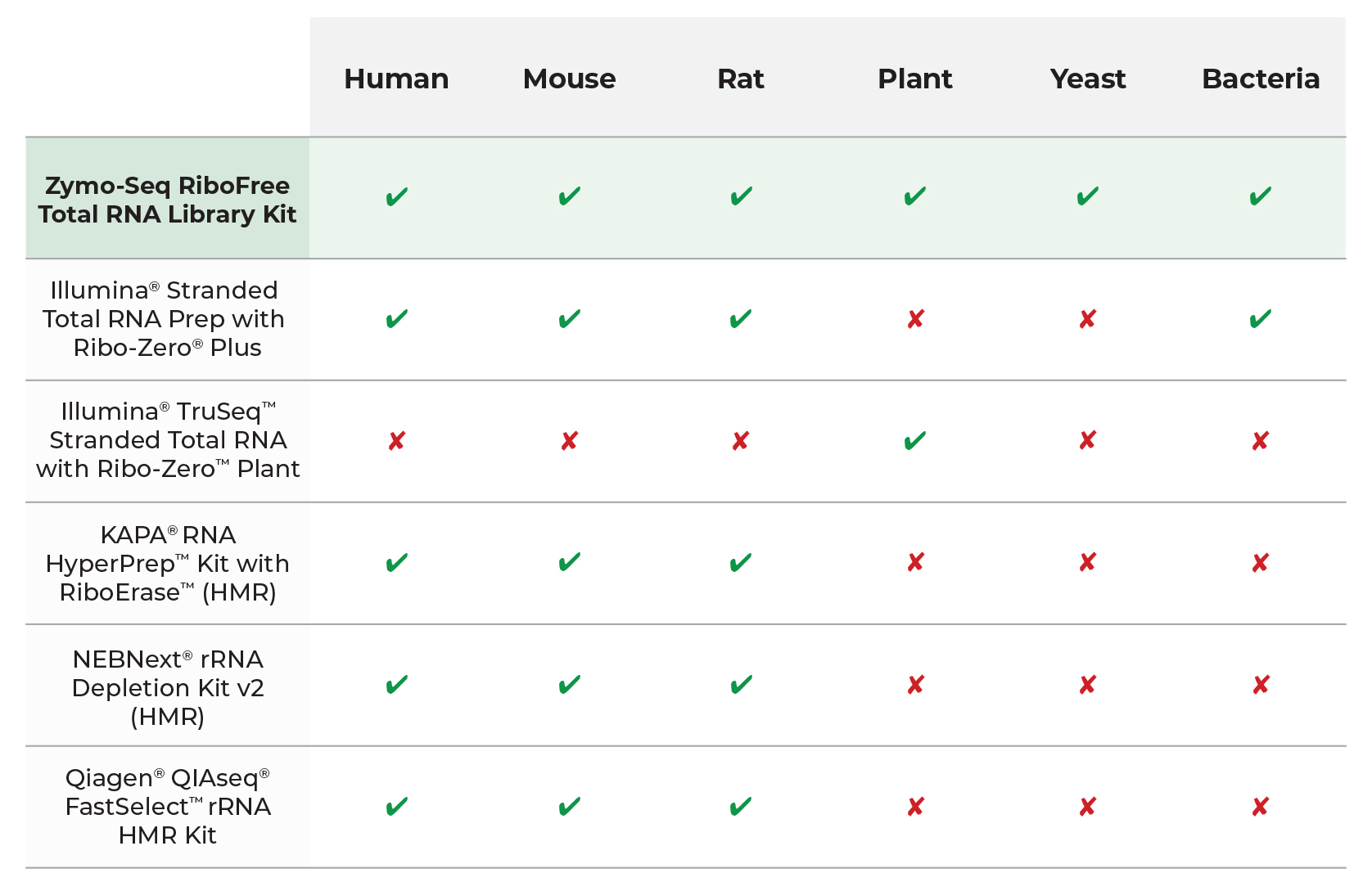
RiboFree Universal Depletion Provides Cross-Species Compatibility. The Zymo-Seq RiboFree Total RNA Library Kit is unmatched in cross-species compatibility thanks to an innovative probe-free depletion strategy. Leveraging the robust depletion kinetics, scientists are exploring diverse applications of RiboFree using samples from vertebrates, plants and more.
Suitable for Degraded Samples*
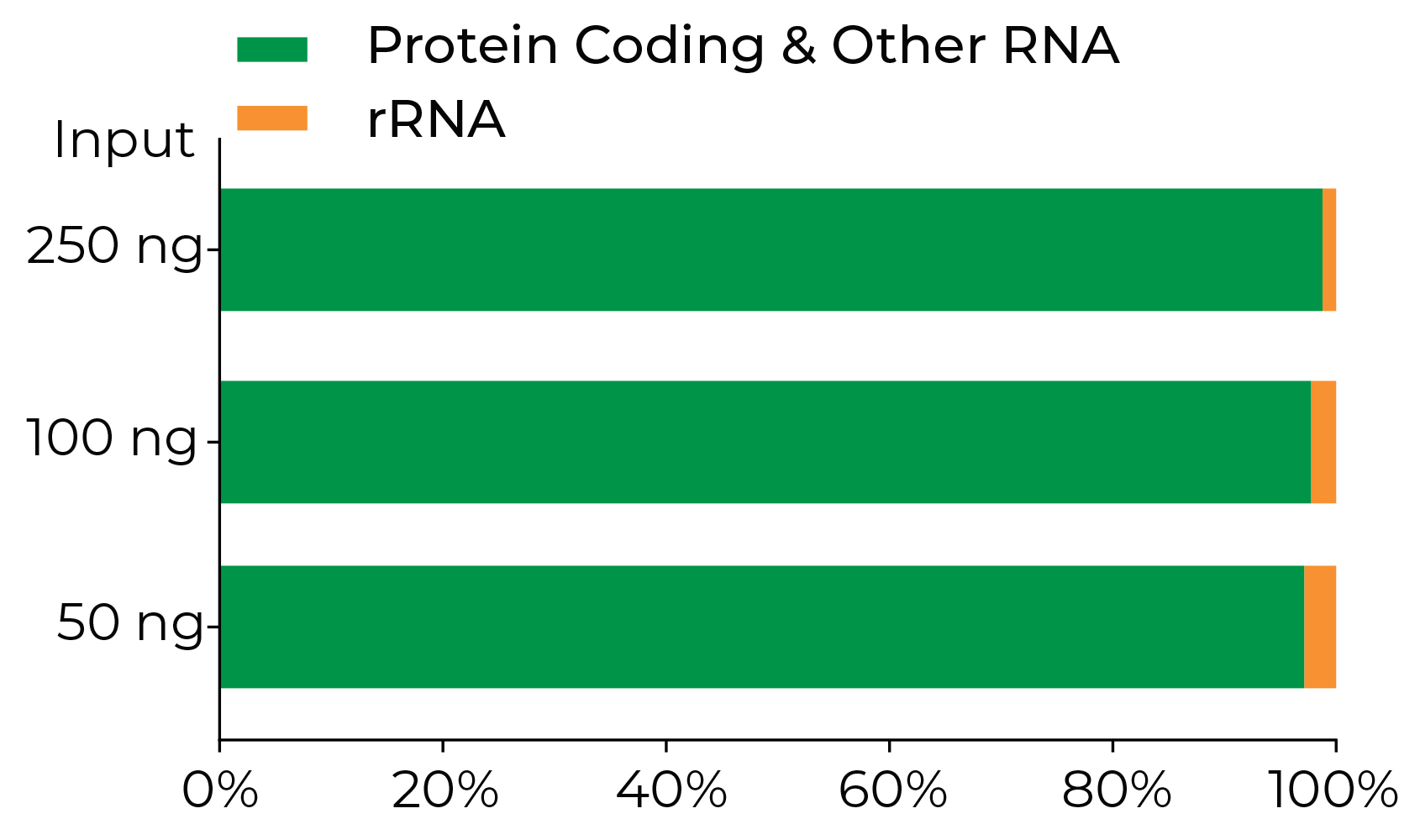
The Zymo-Seq RiboFree Kit Effectively Depletes rRNA from Formalin-Fixed, Paraffin-Embedded (FFPE) Human Tissue RNA. Libraries were prepared with 250 ng, 100 ng, and 50 ng of total RNA extracted from FFPE tissues (RIN = 1.6, peak < 200 nt), respectively, with minimal modifications to the standard protocol as described in Appendix E of the instruction manual.
*Please contact tech@zymoresearch.com for additional recommendations.
The Fastest RNA-Seq Library Workflow
Sample to Sequencer in a Single Day
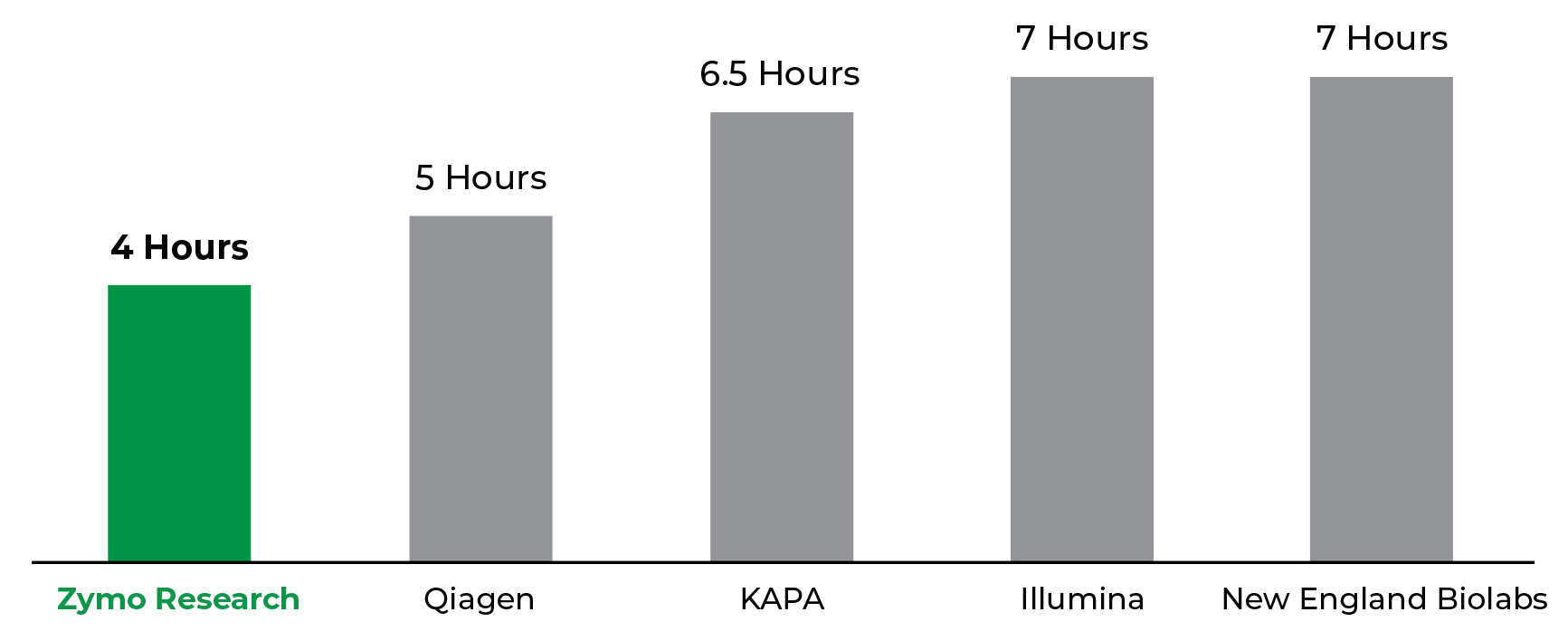
The Zymo-Seq RiboFree Kit has the Fastest Workflow from Total RNA to rRNA-depleted, NGS Libraries. The Zymo-Seq RiboFree Total RNA Library Kit offers a 20% - 40% reduction in workflow completion time compared to other available RNA library prep kits. Workflow time considers the hours required to complete both rRNA depletion and library preparation.
Recent Citations
The RiboFree® technology has seen application across a broad spectrum of research areas such as animal genome annotation, fundamental mechanism elucidation, microbial research/metatranscriptomics, pathogen detection, plant research, and many more. See below for a few recent publications.
- M. Keremane, K. Singh, C. Ramadugu, R. R. Krueger, T. H. Skaggs, Next Generation Sequencing, and Development of a Pipeline as a Tool for the Detection and Discovery of Citrus Pathogens to Facilitate Safer Germplasm Exchange. Plants (Basel) 13, (2024).
- E. Orcel et al., A single workflow for multi-species blood transcriptomics. BMC Genomics 25, 282 (2024).
- M. V. Martín-Manzo, R. M. Morelos-Castro, A. Munguia-Vega, M. L. Soberanes-Yepiz, E. Cortés-Jacinto, Transcriptome analysis of reproductive tract tissues of male river prawn Macrobrachium americanum. Mol Biol Rep 51, 259 (2024).
- P. S. Salazar-Hamm et al., Choclo virus (CHOV) recovered from deep metatranscriptomics of archived frozen tissues in natural history biorepositories. PLoS Negl Trop Dis 18, e0011672 (2024).
- J. Wang et al., Individual bat virome analysis reveals co-infection and spillover among bats and virus zoonotic potential. Nat Commun 14, 4079 (2023).
- T. J. Brine et al., Identification and characterization of Phaseolus vulgaris endornavirus 1, 2 and 3 in common bean cultivars of East Africa. Virus Genes 59, 741-751 (2023).
- G. J. S. Talross, J. R. Carlson, The rich non-coding RNA landscape of the Drosophila antenna. Cell Rep 42, 112482 (2023).
- M. Malone et al., Host-microbe metatranscriptome reveals differences between acute and chronic infections in diabetes-related foot ulcers. APMIS 130, 751-762 (2022).
- E. G. Overbey et al., Transcriptomes of an Array of Chicken Ovary, Intestinal, and Immune Cells and Tissues. Front Genet 12, 664424 (2021).
- J. Gray et al., Colonisation dynamics of Listeria monocytogenes strains isolated from food production environments. Sci Rep 11, 12195 (2021).
- D. Willemse, C. Moodley, S. Mehra, D. Kaushal, Transcriptional Response of Mycobacterium tuberculosis to Cigarette Smoke Condensate. Front Microbiol 12, 744800 (2021).
- J. Santamaría-Gómez et al., Role of a cryptic tRNA gene operon in survival under translational stress. Nucleic Acids Res 49, 8757-8776 (2021).
- P. Kanodia, W. A. Miller, Effects of the noncoding subgenomic RNA of red clover necrotic mosaic virus in virus infection. J Virol, JVI0181521 (2021).
- E. M. Wampande et al., Phylogenetic Characterization of Crimean-Congo Hemorrhagic Fever Virus Detected in African Blue Ticks Feeding on Cattle in a Ugandan Abattoir. Microorganisms 9, (2021).
- K. V. Brussel et al., Identification of Novel Astroviruses in the Gastrointestinal Tract of Domestic Cats. Viruses 12, (2020).
- D. Licastro et al., Isolation and Full-Length Genome Characterization of SARS-CoV-2 from COVID-19 Cases in Northern Italy. J Virol 94, (2020).
- N. K. Sinha et al., EDF1 coordinates cellular responses to ribosome collisions. Elife 9, (2020).
- D. O. Omelchenko et al., Assembly and Analysis of the Complete Mitochondrial Genome of Capsella bursa-pastoris. Plants (Basel) 9, (2020).