Successfully Added to Cart
Customers also bought...
-
 DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the...
DNA/RNA Shield (50 ml)Cat#: R1100-50DNA/RNA Shield reagent is a DNA and RNA stabilization solution for nucleic acids in any biological sample. This DNA and RNA stabilization solution preserves the... -
 DNA/RNA Shield SafeCollect Swab Collection Kit, 1ml (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA Shield completely inactivates harmful pathogens...
DNA/RNA Shield SafeCollect Swab Collection Kit, 1ml (1 collection kit)Cat#: R1160The DNA/RNA Shield SafeCollect Swab Collection Kit is a user-friendly collection kit for stabilizing the nucleic acid content of samples collected with a swab. DNA/RNA Shield completely inactivates harmful pathogens...
Quick-16S NGS Library Prep Kit
| Cat # | Name | Size | Price | Quantity |
|---|
Highlights
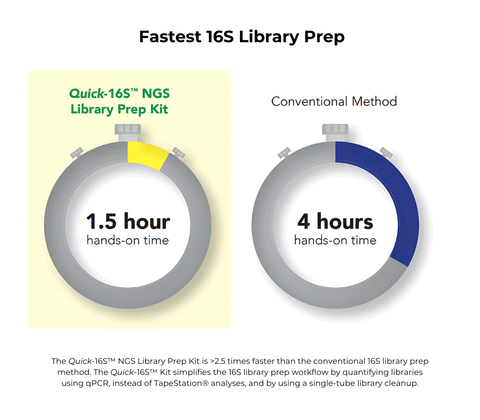
- Fastest: Only 1.5 hours of hands-on time. No Tapestation® analysis or AMPure® clean-ups.
- Accurate: Utilization of real-time PCR limits PCR chimera formation.
- Increased Coverage: Novel primers increase phylogenetic coverage of Bacteria and Archaea and enable species-level resolution for human microbiome profiling.
Documents
Product Description
Technical Specifications
| Amplicon Size | The size of the 16S V1-V2 region and the 16S V3-V4 region (including primers) is ~350bp and ~460bp, respectively. The final amplicon size after addition of barcoded primers is ~486bp and ~596bp, respectively. |
|---|---|
| Barcode Sequences | Available here |
| Equipment | Microcentrifuge, plate spinner (centrifuge), 96-well real-time quantitative PCR system, 96-well real-time PCR plates. |
| Index Primers | Dual index (barcodes) to uniquely label 96 samples. |
| Preparing > 96 Samples | For guidelines to preparing more than 96 samples, please click here. |
| Recommended RT PCR System | Bio-Rad CFX96 Real-Time PCR Detection System (any model), Applied Biosystems 7500 Real-Time PCR System. |
| Sample Input | Purified microbial DNA ≤ 20 ng/µl, free of PCR inhibitors. |
| Sequencing Compatibility | Illumina MiSeq without the need to add custom sequencing primers. Zymo Research recommends the MiSeq Reagent Kit v3 (600-cycle). |
Resources
“This is a very good kit for library preparation, especially compared to other library preparation products. We love that all the reagents we need to prepare a targeted sequencing library are included!”
- Leyao W. (Washington University in St. Louis)
"We first tested this kit during the beta test in 2018. We really liked it an immediately ordered full kits to change our workflow."
- Lisa D. (University of Hohenheim)
"This is an excellent kit and I highly recommend it."
- Karina S. (Universidad Nacional de Misiones)
The authors investigated the microbial pools associated with landfill reactors that are impacted by nanomaterials. By preparing 16S rRNA libraries with the Quick-16S NGS Library Prep Kit, they discovered that the bacterial communities were similar between control reactors and nanomaterial-containing reactors, but the archaea communities were highly different.
Akyol, Ç., Ozbayram, E.G., Demirel, B., Onay, T.T., Ince, O., and Ince, B. (2019). Linking nano-ZnO contamination to microbial community profiling in sanitary landfill simulations. Environ Sci Pollut Res. doi: 10.1007/s11356-019-04906-8This study investigated how varying interchange ratios in wastewater treatment affects microbial community composition. Analysis of samples prepared for 16S sequencing with the Quick-16S NGS Library Prep Kit demonstrated that the relative abundance of the phyla Proteobacteria, Acidobacteria, and Bacteroidetes varied according to the interchange ratio and that the species of Thiothrix were highly abundance in all samples, revealing their potential importance to wastewater treatment processes.
Karlikanovaite-Balikci, A., Ozbayram, E.G., Yagci, N., and Ince, O. (2019). Microbial community shifts in the oxic-settling-anoxic process in response to changes to sludge interchange ratio. Heliyon, 5(4), e01517. doi: 10.1016/j.heliyon.2019.e01517Akyol et al. performed 16S and 18S analysis of anaerobic digestates using the Quick-16S NGS Library Prep Kit to assess microbial diversity in lignocellulose-rich digestates. The most abundant bacterial and fungal genera were consistent across digestate samples.
Akyol, Ç., Ince, O., and Ince, B. (2019). Crop-based composting of lignocellulosic digestates: Focus on bacterial and fungal diversity. Bioresour. Technol., 121549. doi: 10.1016/j.biortech.2019.121549Read More
Need help? Contact Us